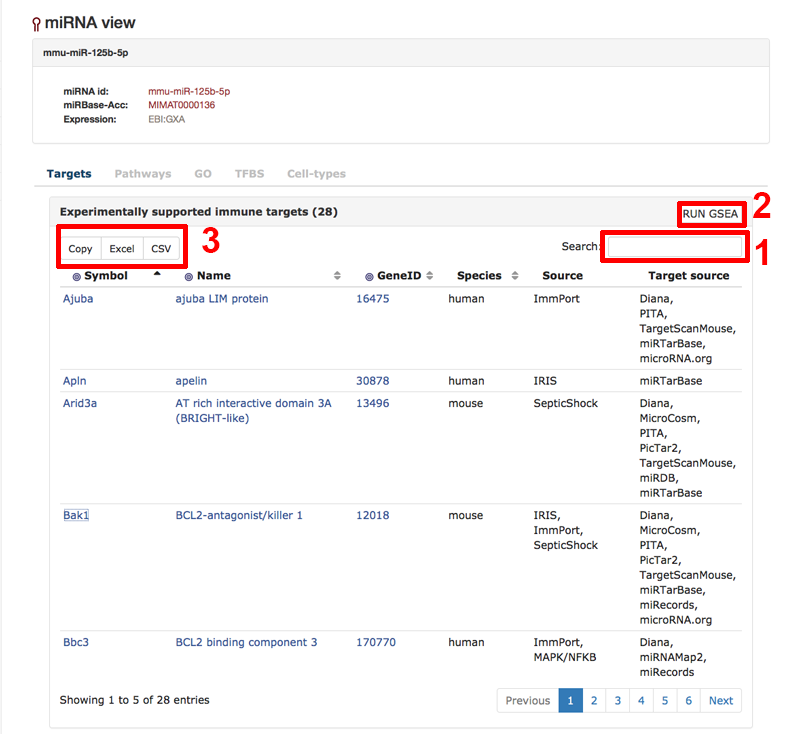
Here, we have look at the miRNA view of an individual miRNA found e.g. through the browse/search function or through a target view of the site. In this example, we look at the miRNA mmu-miR-125b-5p. The first image below shows the "Targets"-tab of the miRNA view. Here one can investigate the experimentally supported and predicted targets of the miRNA. The table can be searched using the search field (1). One can use all targets of the table to run a gene set enrichment analysis via Enrichr [Kuleshov et al.] by clicking on the button in the right hand corner of the table (2). One can also download the content of the table (3).

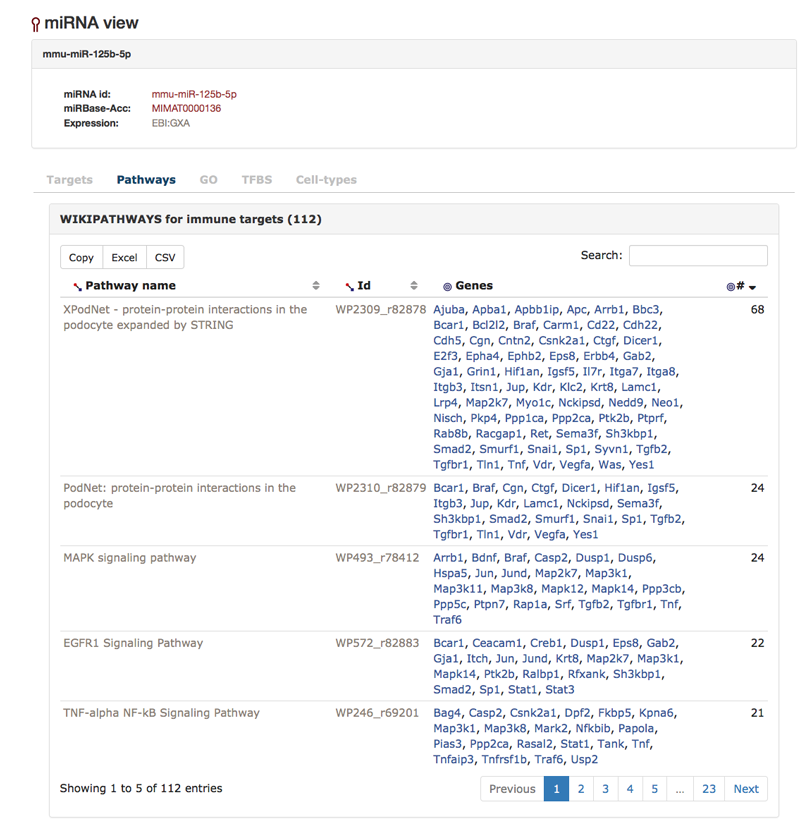
The next tab is the "Pathways"-tab of the miRNA view. Here one can investigate the pathways that the targets of the miRNA are part of. Currently, IRNdb holds data for KEGG, Wikipathway and Reactome.
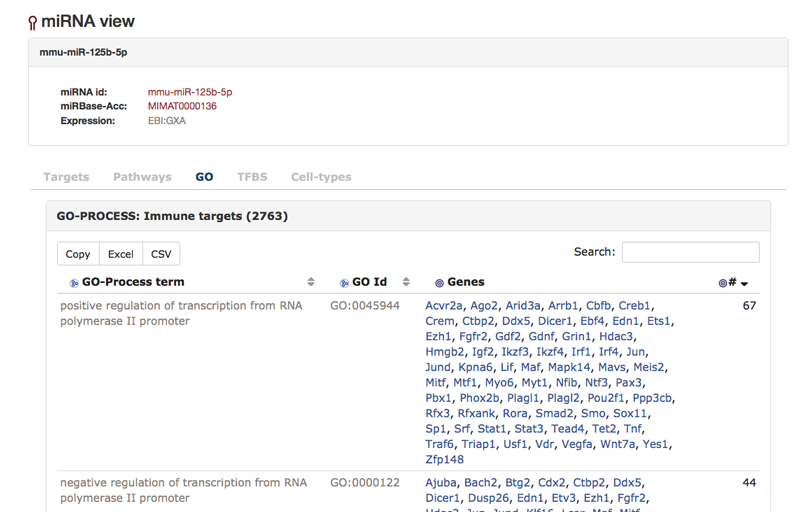
On the "GO"-tab of the miRNA view one can investigate the Gene Ontology terms that the targets of the miRNA are associated with.
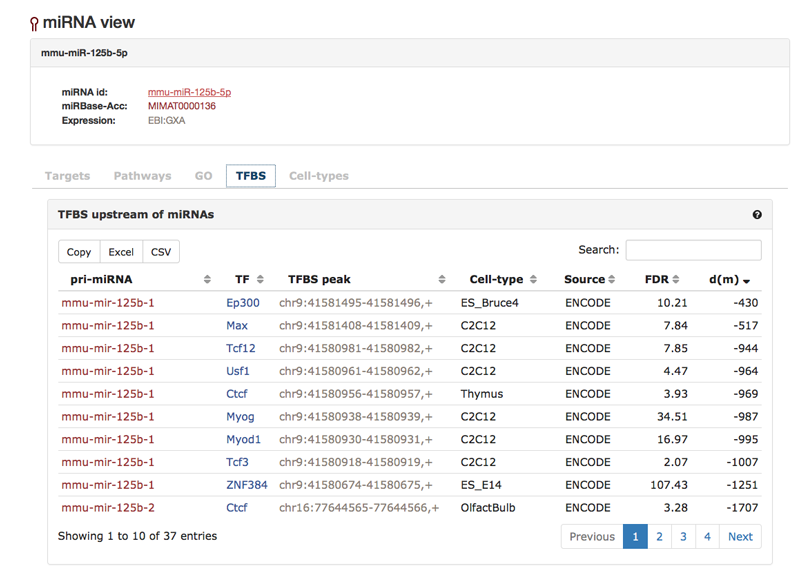
The "TFBS"-tab shows transcription factor binding sites based on ChIP experiments that we collected. We extracted pri-miRNAs (based on miRBase) for the mature miRNAs. Next, we mapped within 2500nt upstream of the 5'end of the pir-miRNA ChIP-seq data from several sources (see "Resources") based on the GRCm38 genome build. A mature miRNA can have more than one pri-miRNA, thus the table may contain more than one pri-miRNA for one mature miRNA under investigation. The table list the pri-miRNAs as well as the TFs that could be mapped in their vicinity. We also show the cell-type in which the ChIP-seq experiment was performed, the FDR with which the ChIP-seq region could be mapped as well as the distance of the ChIP-seq region to the 5'end of the pri-miRNA.
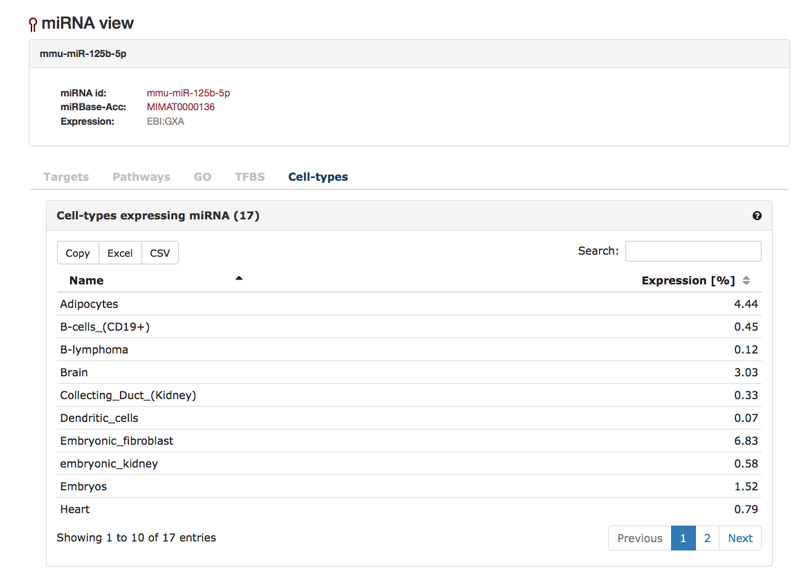
Finally, the "Cell-types"-tab shows the expression of the miRNA in different cell-types based on a miRNA expression atlas [Landgraf et al.]. The percentage shows the miRNA contribution in expression on the total expression of all miRNAs expressed in the cell-type.
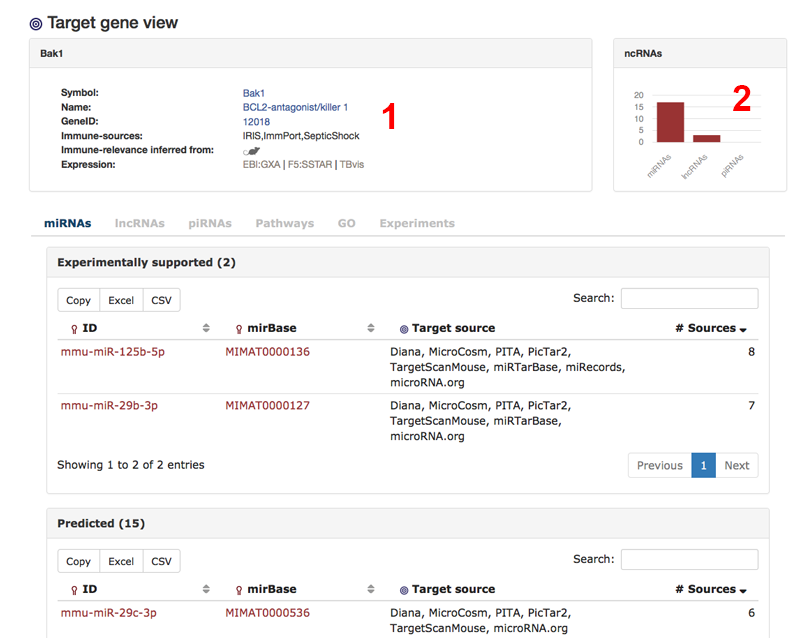
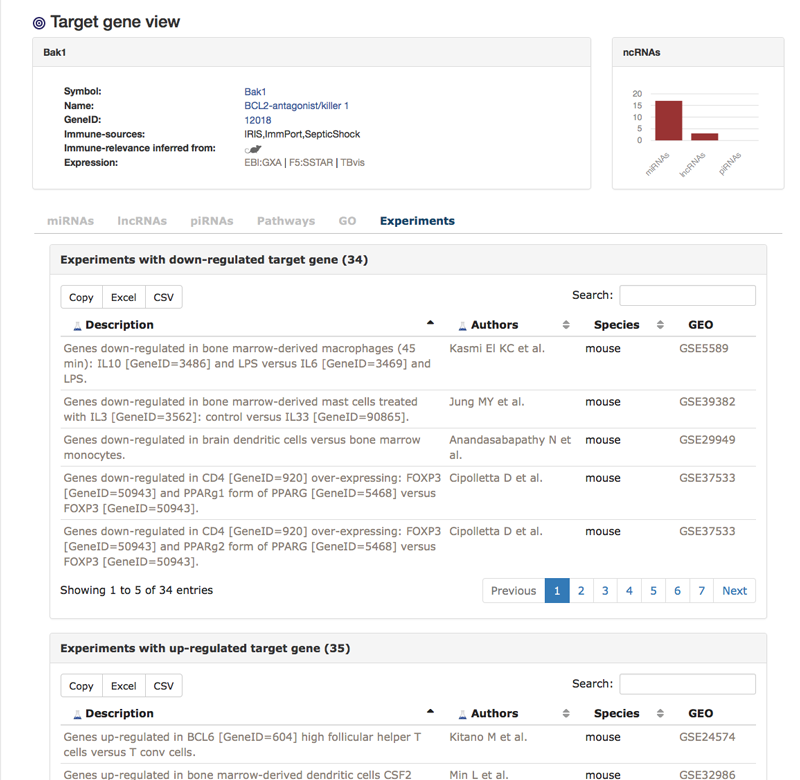
This is the view for one particular target gene. In this example we are looking at target gene Bak1. There is some general information about the gene (1) as well as a quick overview of associated ncRNAs (2). The first tab shows all miRNAs that are either confirmed or predicted to be targeting Bak1. We also are able to see the sources.
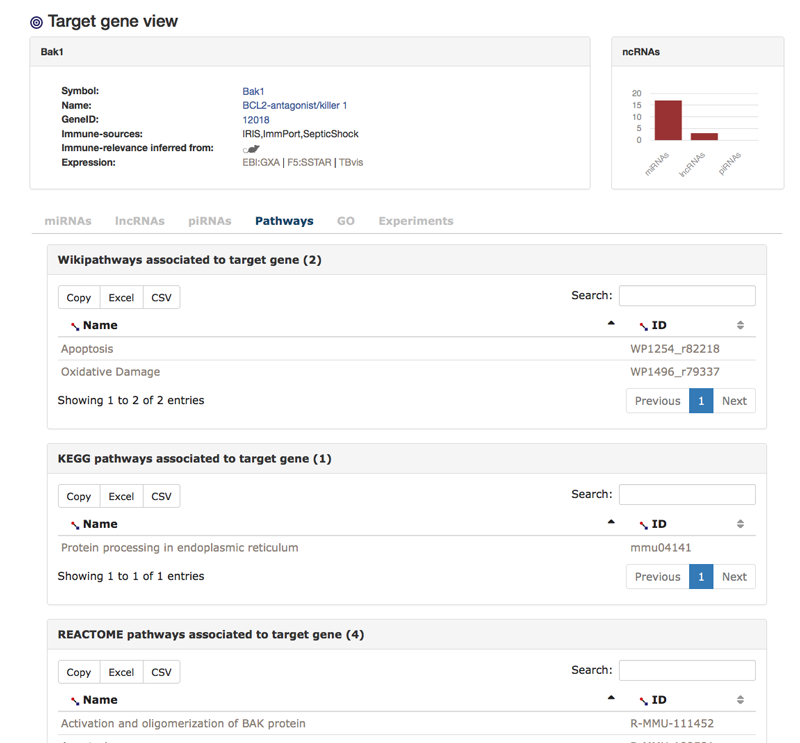
The next two tabs are similar to the miRNA views tabs but for piRNAs and lncRNAs that interact with the target gene. The next two tabs show pathway and GO-terms that are associated with the target gene. In the pathway view, one can find links to IRNdb individual pathway view pages that show all targets of ncRNAs in the pathway (see below "The pathway view").
The "GO"-tab shows all GO-terms that are associated to the target gene.
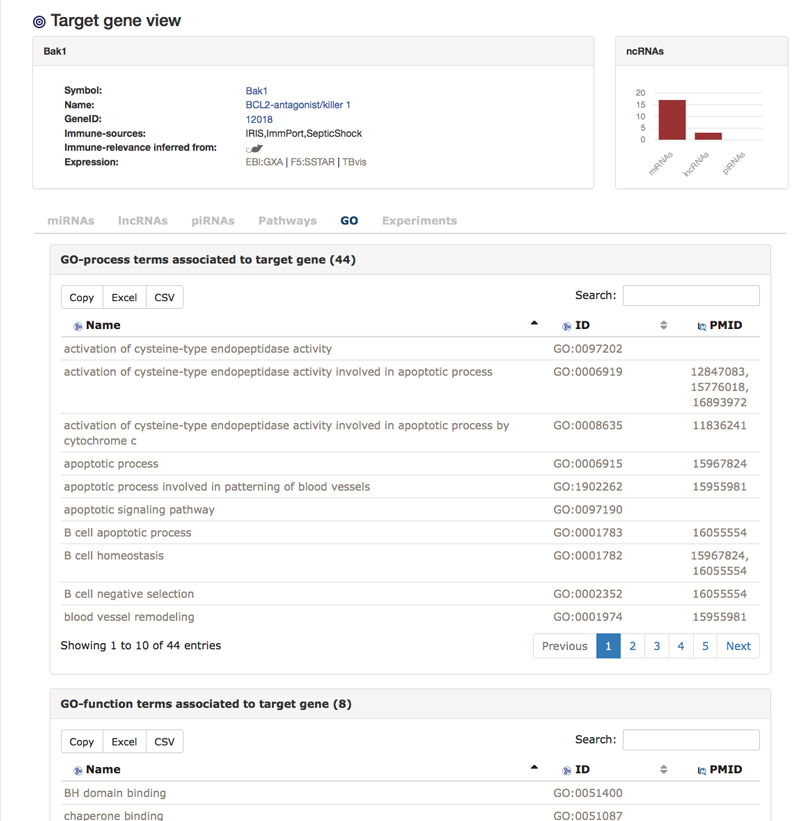
The "Experiments"-tab shows immunologically themed experiments in which the target gene is either down- or up-regulated. We also find a link to MSigDB [Liberzon A et al.] where the information was extracted from and a link to the original publication and dataset if available.
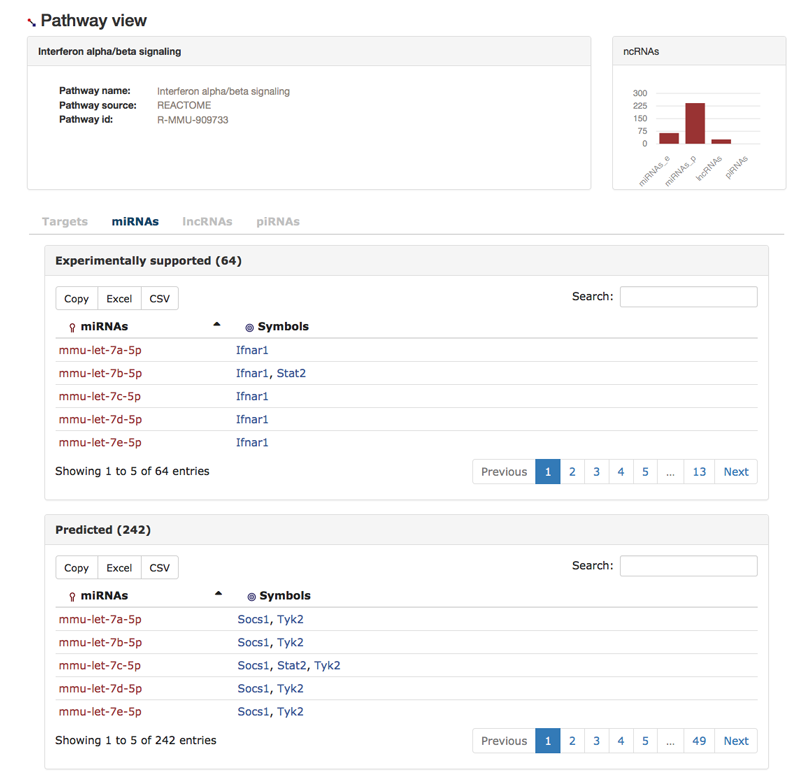
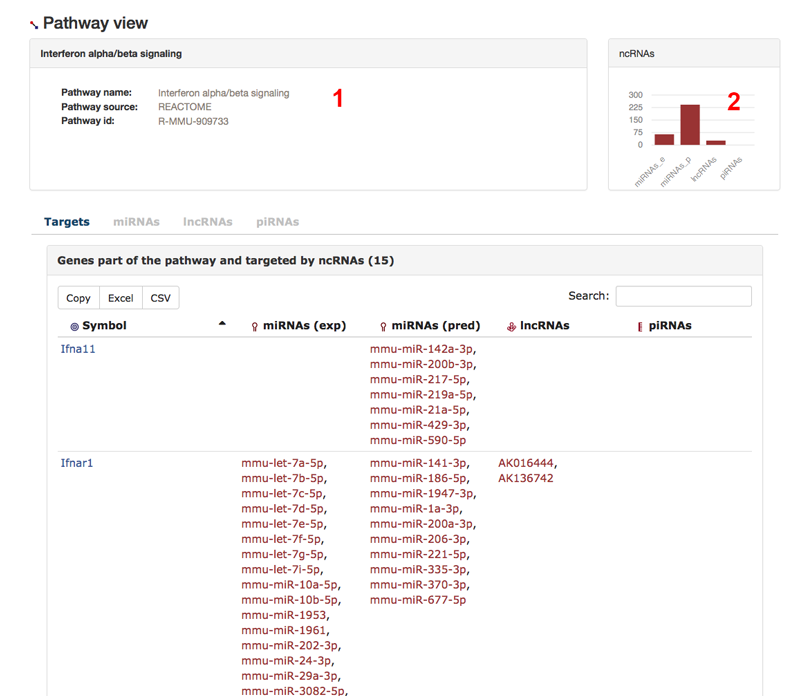
Here, we have look at the pathway view. In this example, we look at the REACTOME pathway Interferon alpha/beta signaling. We find some general information about the pathway (1) and the numbers of miRNAs, lncRNAs, and piRNA that target genes in the pathway (2). We present tables that show the elements associated to the pathway from the view of different biological entities, e.g. the 'Targets'-tab table shows for each ncRNA-targeted gene in the pathway the gene and its interacting ncRNAs.
Finally, we show in the ncRNA-tabs the same information from the view-point of the ncRNA type in question, e.g. the next image shows the 'miRNAs'-tab.